The 2024 Nobel Prize in Chemistry: AI Revolutionizes Protein Structure Prediction

In a groundbreaking announcement that marks a new era in the intersection of artificial intelligence and biochemistry, the Royal Swedish Academy of Sciences has awarded the 2024 Nobel Prize in Chemistry to three pioneering researchers for their revolutionary work in AI-driven protein structure prediction. This year's laureates, Demis Hassabis, John Jumper, and David Baker, have fundamentally transformed our understanding of protein folding and structure, opening up new frontiers in drug discovery, disease treatment, and biotechnology.
1. Introduction: A Breakthrough Year for AI in Chemistry
The decision to award the Nobel Prize in Chemistry to researchers in the field of AI-driven protein structure prediction underscores the growing importance of artificial intelligence in scientific discovery. This recognition comes at a pivotal moment when the power of AI to solve complex scientific problems has become undeniable.
Proteins, the building blocks of life, perform a vast array of functions in living organisms, from catalyzing chemical reactions to providing structural support for cells. The function of a protein is intimately tied to its three-dimensional structure, which is determined by the way its chain of amino acids folds into a complex shape. Understanding and predicting these structures has been one of the grand challenges in biology for decades, known as the "protein folding problem."
Traditional methods of determining protein structures, such as X-ray crystallography and cryo-electron microscopy, while powerful, are time-consuming, expensive, and not always successful. The ability to accurately predict protein structures from their amino acid sequences using computational methods has been a long-standing goal in the field of structural biology.
The work of this year's Nobel laureates represents a quantum leap in our ability to predict protein structures with unprecedented accuracy using artificial intelligence. Their achievements have not only solved a fundamental problem in biology but have also paved the way for rapid advancements in drug discovery, understanding disease mechanisms, and designing novel proteins with specific functions.
2. The Laureates: Pioneers in AI-Driven Protein Structure Prediction
Demis Hassabis: The Visionary Behind DeepMind and AlphaFold
Demis Hassabis, co-founder and CEO of DeepMind, has been at the forefront of artificial intelligence research for over two decades. Born in London in 1976, Hassabis was a child prodigy in chess, reaching the rank of master at the age of 13. He completed his A-levels at the age of 16 before going on to graduate from Cambridge University with a double first in Computer Science.
Hassabis's journey in AI began long before the founding of DeepMind. His PhD in cognitive neuroscience from University College London focused on memory and imagination, laying the groundwork for his later work in AI. In 2010, he co-founded DeepMind with the ambitious goal of creating artificial general intelligence (AGI) - AI systems capable of performing any intellectual task that a human can.
Under Hassabis's leadership, DeepMind has achieved numerous breakthroughs in AI, including the development of AlphaGo, the first computer program to defeat a world champion at the game of Go. However, it is the company's work on AlphaFold that has earned Hassabis his share of the Nobel Prize.
Hassabis's vision for AI extends far beyond games and into the realm of scientific discovery. He has long advocated for the use of AI to accelerate scientific progress, particularly in areas that have proven challenging for traditional methods. The success of AlphaFold in predicting protein structures with unprecedented accuracy is a testament to this vision.
John Jumper: Lead Researcher of the AlphaFold Project
John Jumper, a senior research scientist at DeepMind, has been the driving force behind the development of AlphaFold. With a background in physics and a PhD from the University of Chicago, Jumper brought a unique perspective to the problem of protein structure prediction.
Jumper's work on AlphaFold began in 2016 when DeepMind decided to tackle the protein folding problem. Under his leadership, the AlphaFold team made rapid progress, culminating in their groundbreaking performance at the 14th Critical Assessment of Protein Structure Prediction (CASP) competition in 2020.
Jumper's approach to the problem was characterized by a deep understanding of both the biological aspects of protein folding and the potential of deep learning algorithms. He and his team developed novel neural network architectures that could effectively learn the complex patterns in protein structures from existing data.
One of Jumper's key insights was the importance of incorporating evolutionary information into the AI model. By analyzing the patterns of amino acid substitutions in related proteins across different species, AlphaFold could infer important constraints on the protein's structure, greatly improving its predictions.
David Baker: The Protein Design Pioneer
David Baker, a professor of biochemistry at the University of Washington and director of the Institute for Protein Design, has been a leading figure in the field of protein structure prediction and design for over two decades. His work has been instrumental in advancing our understanding of protein folding and in developing methods for designing novel proteins with specific functions.
Baker's journey in protein structure prediction began in the 1990s with the development of the Rosetta software suite. Rosetta uses a combination of physical principles and statistical analysis to predict and design protein structures. Over the years, Baker and his team have continually refined and expanded Rosetta's capabilities, making it one of the most powerful tools in computational structural biology.
In recent years, Baker has been at the forefront of integrating machine learning techniques into protein structure prediction and design. His lab's development of the RoseTTAFold algorithm, which combines deep learning with traditional physics-based approaches, has achieved results comparable to AlphaFold in many cases.
Baker's work extends beyond prediction to the de novo design of proteins with specific functions. His lab has created novel proteins that can catalyze chemical reactions, serve as potential vaccines, or act as molecular switches. This ability to design proteins from scratch opens up enormous possibilities in biotechnology and medicine.
3. AlphaFold: The Game-Changing AI System
AlphaFold, developed by DeepMind, represents a quantum leap in the field of protein structure prediction. Its ability to predict protein structures with atomic-level accuracy has been hailed as a solution to the 50-year-old protein folding problem.
What is AlphaFold and How Does it Work?
At its core, AlphaFold is a deep learning system that takes a protein's amino acid sequence as input and predicts its three-dimensional structure. The system uses a novel neural network architecture that combines attention mechanisms, similar to those used in natural language processing, with algorithms designed to capture the geometric constraints of protein structures.
One of the key innovations in AlphaFold is its use of multiple sequence alignments (MSAs) as input. By analyzing the patterns of amino acid substitutions in related proteins across different species, AlphaFold can infer important constraints on the protein's structure. This evolutionary information is combined with physical and geometric constraints to guide the prediction process.
AlphaFold also employs a novel approach called "iterative refinement." The system makes an initial prediction of the protein's structure, then repeatedly refines this prediction, each time focusing on different aspects of the structure. This process allows AlphaFold to gradually improve its predictions, often resulting in highly accurate final structures.
The Evolution from AlphaFold to AlphaFold 3
Since its initial release, AlphaFold has undergone significant improvements. AlphaFold 2, released in 2020, represented a major leap forward in accuracy, achieving unprecedented performance in the CASP14 competition. It was able to predict structures with an average Global Distance Test (GDT) score of 92.4 out of 100, meaning its predictions were typically off by less than the width of an atom.
The latest version, AlphaFold 3, released in 2023, further extends the capabilities of the system. It can now predict not only the structures of individual proteins but also how proteins interact with other molecules, including other proteins, DNA, RNA, and small molecule ligands. This ability to model protein complexes and interactions opens up new possibilities for understanding cellular processes and designing drugs.
Comparison with Traditional Methods of Protein Structure Prediction
Traditional methods of protein structure determination, such as X-ray crystallography and cryo-electron microscopy, while highly accurate, are time-consuming and expensive. They also require the protein to be isolated and often crystallized, which is not always possible for all proteins.
Computational methods for protein structure prediction have been in development for decades, but their accuracy has been limited. Before AlphaFold, the best computational methods could typically predict the overall fold of a protein correctly but often struggled with the details of the structure, particularly for proteins with no close relatives of known structure.
AlphaFold represents a step-change in the accuracy of computational prediction. It can often produce predictions that are as accurate as experimental structures, and it can do so for a wide range of proteins, including those with no known structural relatives. Moreover, AlphaFold can produce these predictions in a matter of hours or days, compared to the months or years often required for experimental structure determination.
This combination of speed and accuracy makes AlphaFold a game-changing tool in structural biology. It allows researchers to rapidly generate structural models for proteins that have been resistant to experimental methods, opening up new avenues for research in areas ranging from basic biology to drug discovery.
4. Impact on Biological Sciences and Medicine
The ability to accurately predict protein structures has far-reaching implications across biology and medicine. AlphaFold and related AI systems are already having a significant impact in several key areas:
Accelerating Drug Discovery and Development
One of the most immediate and significant impacts of AI-driven protein structure prediction is in the field of drug discovery. The structure of a protein is crucial for understanding how it might interact with potential drug molecules. With accurate structural predictions, researchers can:
- Identify potential binding sites on proteins that could be targeted by drugs.
- Design drugs that are more likely to bind effectively to their target proteins.
- Predict potential off-target interactions that could lead to side effects.
- Understand how mutations in proteins might affect drug binding, helping to predict drug resistance.
Several pharmaceutical companies have already begun integrating AlphaFold predictions into their drug discovery pipelines. For example, researchers at the University of Cambridge used AlphaFold to identify a potential new drug target for malaria parasites.
Advancing Understanding of Genetic Diseases
Many genetic diseases are caused by mutations that affect protein structure and function. With AI-driven structure prediction, researchers can now:
- Predict how disease-causing mutations might affect protein structure.
- Identify potential compensatory mutations that could restore protein function.
- Understand the structural basis of protein misfolding diseases like Alzheimer's and Parkinson's.
For instance, researchers have used AlphaFold to gain insights into the structure of the BRCA1 protein, mutations in which are associated with increased risk of breast and ovarian cancer. These structural insights could help in developing new treatments or in interpreting genetic test results.
Potential Applications in Vaccine Design
The COVID-19 pandemic has highlighted the importance of rapid vaccine development. AI-driven protein structure prediction could accelerate vaccine design in several ways:
- Predicting the structures of viral proteins, helping to identify potential vaccine targets.
- Designing stable and immunogenic protein antigens for use in vaccines.
- Understanding how mutations in viral proteins might affect vaccine efficacy.
Researchers have already used AlphaFold to study the structures of SARS-CoV-2 proteins, providing insights that could be valuable for future vaccine development.
5. Tencent RTC and the Future of AI in Scientific Education and Communication
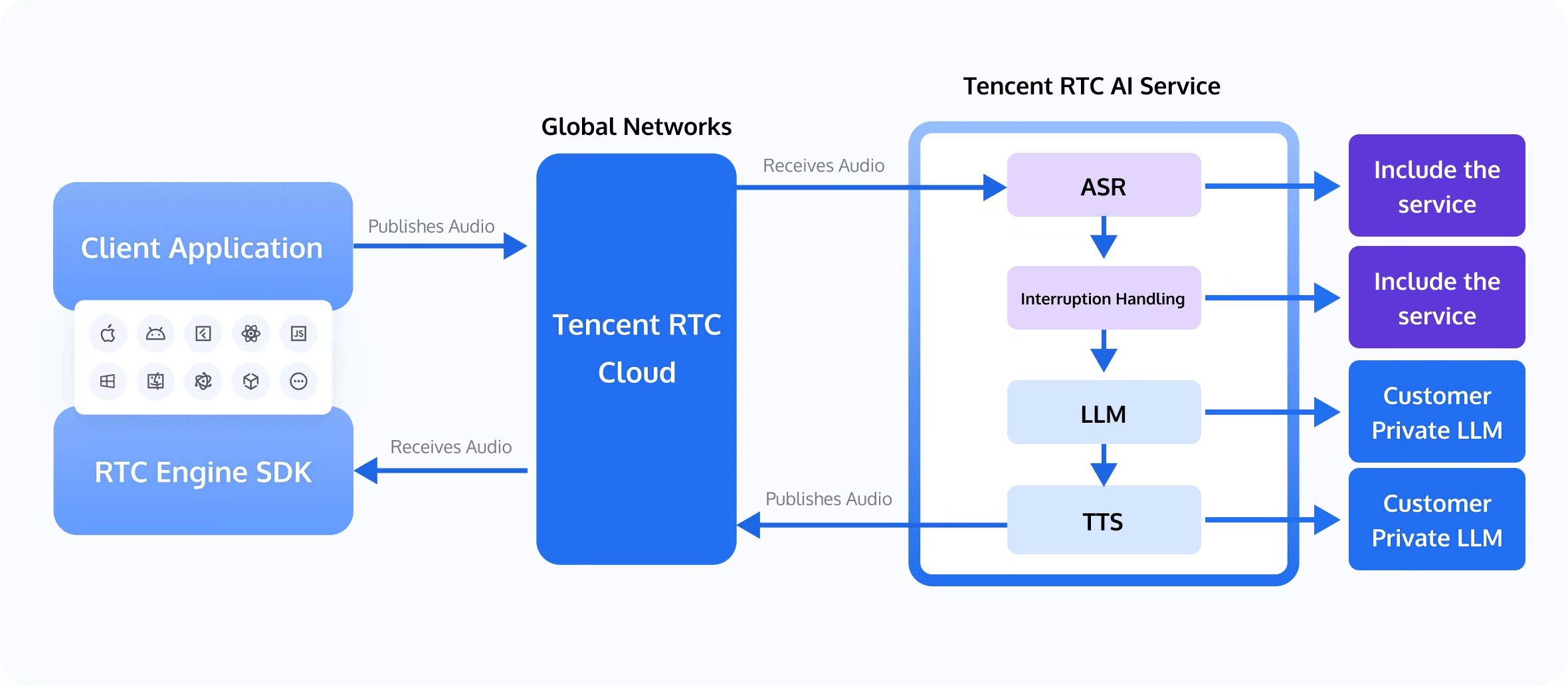
The groundbreaking work in AI-driven protein structure prediction opens up new possibilities for scientific education and communication. Tencent RTC Conversational AI Solutions are at the forefront of this revolution, offering innovative ways to disseminate complex scientific knowledge.
Tencent RTC Conversational AI Solutions combine real-time communication technology with advanced artificial intelligence to create seamless, interactive experiences. These solutions leverage Automatic Speech Recognition (ASR), Text-to-Speech (TTS), Large Language Models (LLM), and Retrieval-Augmented Generation (RAG) to enable natural, intelligent conversations across various applications.
Use conversational AI to provide a more personalized educational experience
- AI Speaking Coach
- AI Virtual Teacher
- Character Dialogue
- Scenario Practice
Integrating voice capabilities allows for the creation of virtual teaching assistants that mimic real-time human interaction, providing personalized instruction and responsive feedback within educational scenarios.
Elevate social interactions and entertainment experiences with conversational AI
- Virtual AI Companion
- Character AI Dialogue
- Interactive Game
- Metaverse
Utilizing conversational AI combined with real-time interaction capabilities to understand user intentions and provide corresponding feedback, delivering a more realistic and personalized social entertainment experience for users.
Streamline workflows and boost efficiency with conversational AI
- Voice Search Assistant
- Voice Translation Assistant
- Schedule Assistant
- Office Assistant
Voice-activated productivity tools enable users to command and control applications with their voice, increasing efficiency and reducing manual input.
Elevate call center operations with conversational AI
- AI Customer Service
- AI Sales Consultant
- Intelligent Outbound Calling
- E-commerce Assistant
Conversational AI in call centers, powered by RAG and voice interaction, provides a rich, real-time customer service experience. It reduces costs and enhances service efficiency.
Why Choose Tencent RTC Conversational AI Solutions?
- Achieve Natural Dialogue with AI: Achieve Natural Dialogue with AI ASR + TTS technology ensures clear speech recognition and precise text-to-speech output, with a variety of voice options for a personalized communication experience.
- Ultra-low Latency Communication: Ultra-low Latency Communication Global end-to-end latency for voice and video transmission between the model and users is less than 300ms, ensuring smooth and uninterrupted communication.
- Deliver Precise and Stable Conversational AI: Deliver Precise and Stable Conversational AI LLM + RAG integration allows users to upload their knowledge bases to reduce misinformation and achieve more targeted and stable conversational AI.
- Emotional Communication Experience: Emotional Communication Experience Sentiment analysis and interruption handling accurately recognize and respond to user emotions, providing an emotionally rich communication experience.
6. Conclusion: A New Era of AI-Powered Scientific Discovery
The awarding of the 2024 Nobel Prize in Chemistry to Hassabis, Jumper, and Baker marks a watershed moment in the history of science. It represents not just a recognition of their groundbreaking work in protein structure prediction, but also an acknowledgment of the transformative potential of artificial intelligence in scientific discovery.
The success of AlphaFold and related AI systems in solving the protein folding problem demonstrates the power of interdisciplinary approaches that combine deep learning with domain-specific knowledge. It shows how AI can complement and accelerate traditional scientific methods, opening up new avenues of research and discovery.
Looking forward, the impact of this work is likely to be felt far beyond the field of structural biology. The techniques developed for protein structure prediction could potentially be adapted to other complex prediction problems in chemistry and biology. Moreover, the success of AI in this domain is likely to inspire increased investment and research into AI applications across all areas of science.
However, as we celebrate these achievements, it's important to remember that AI is a tool that augments human intelligence rather than replacing it. The true power of AI in science comes from its ability to work in partnership with human researchers, combining the pattern-recognition capabilities of machine learning with human creativity and insight.
The work of Hassabis, Jumper, and Baker, and the AI systems they've developed, have given us powerful new tools for understanding the molecular basis of life. As we look to the future, we can anticipate a new era of scientific discovery, where AI and human intelligence work hand in hand to tackle some of the most challenging problems in science and medicine.
From accelerating drug discovery to unraveling the mysteries of protein misfolding diseases, from designing new enzymes for industrial applications to developing next-generation vaccines, the possibilities opened up by AI-driven protein structure prediction are vast and exciting.
As we stand on the brink of this new era, we can look forward to a future where the boundaries of scientific knowledge are pushed ever further, driven by the synergy between human creativity and artificial intelligence. The 2024 Nobel Prize in Chemistry may well be remembered as a pivotal moment in this journey, marking the beginning of a new chapter in the story of scientific discovery.